Spotlight - AG Oehme
Drug sensitivity profiling in the pediatric precision oncology program INFORM
The international multicenter INdividualized therapy FOr high-Risk childhood Malignancies (INFORM) pediatric precision oncology program addresses an unmet medical need by providing comprehensive molecular next generation sequencing information (NGS) including low-coverage whole-genome sequencing, whole-exome sequencing, RNA sequencing, and 850k DNA methylation profiling for relapsed/refractory pediatric cancer patients.
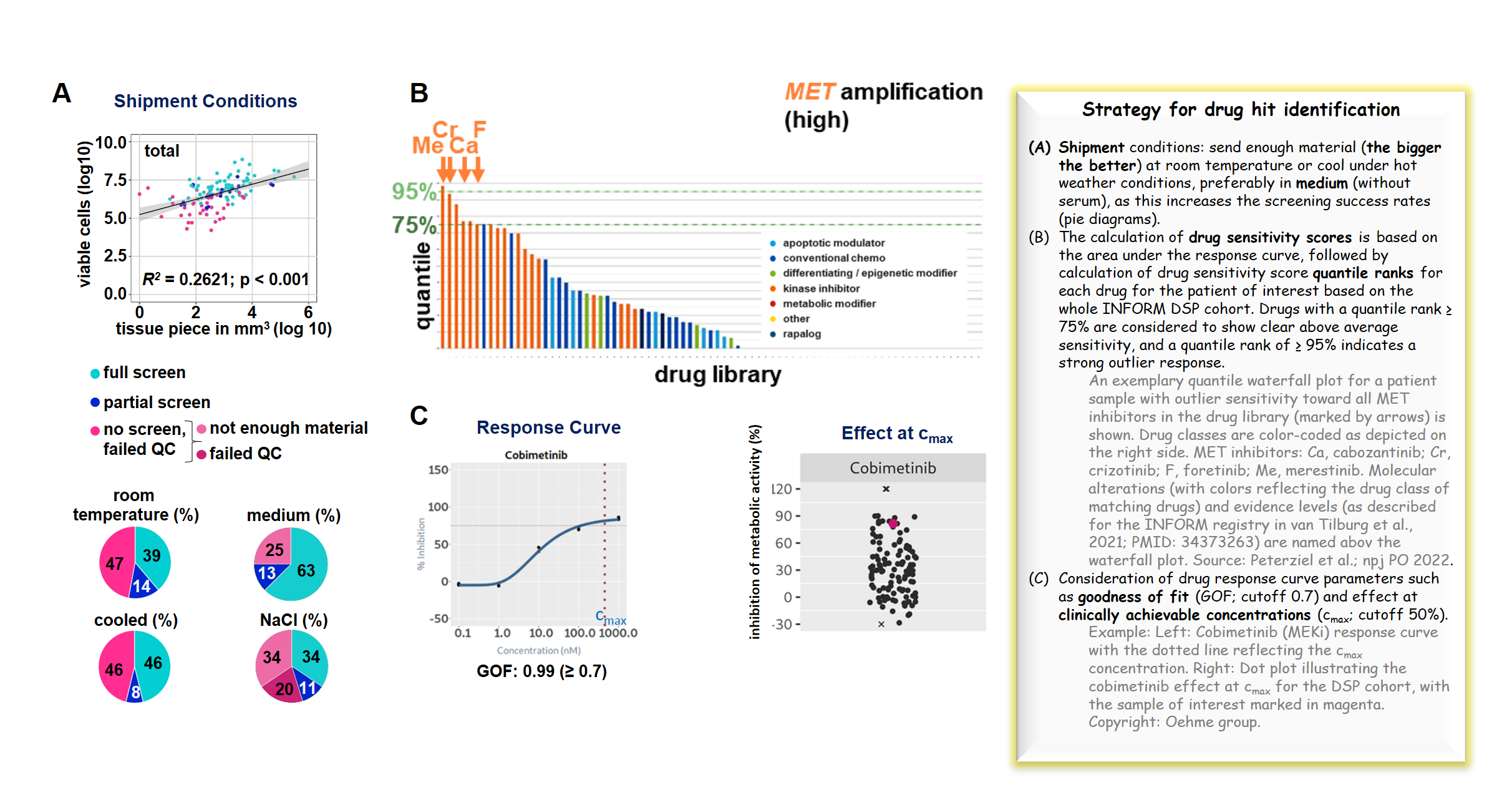
To add a further functional component to INFORM, we initiated a personalized drug sensitivity profiling (DSP) platform based on metabolic activity measurements complementing NGS-based target identification. We have recently published the results of a proof-of-concept two-year pilot study implementing ex vivo DSP on fresh tumor specimens obtained through INFORM (Peterziel et al. 2022, NPJ Precision Oncology, PMID 36575299). We established protocols for diagnosis-specific sample handling, drug screening with a library of clinically relevant drugs, and DSP analysis. Drug screening was performed on patient-derived 3D multicellular fresh tissue cultures within a short time (in most cases two to seven days) after sample acceptance. With some random cases, we confirmed that these cultures still largely reflected the multicellular composition of the original tumor. The submitted material was of sufficient quantity and quality in the majority of the cases to report DSP findings with a turnaround time of approximately three weeks. The DSP results matched the identified molecular targets, including BRAF, ALK, MET, and TP53 status. This was especially evident when including outlier sensitivity detection (within the DSP cohort) in the drug hit selection approach. Adding value to the molecular data, drug vulnerabilities were moreover identified in the majority of cases lacking actionable (very) high-evidence molecular events. With the aim of advancing expanded drug testing approaches and clinical trial development, when applicable, we are establishing organoid-like long-term cell cultures and, in collaboration with other KiTZ groups, murine patient-derived xenograft (mPDX) models of the submitted INFORM fresh tissue samples.
Overall, our proof-of-concept pilot study proved that DSP is feasible within a clinically relevant time and adds valuable functional information when included in precision oncology programs such as INFORM.
Zebrafish xenograft models for drug screening
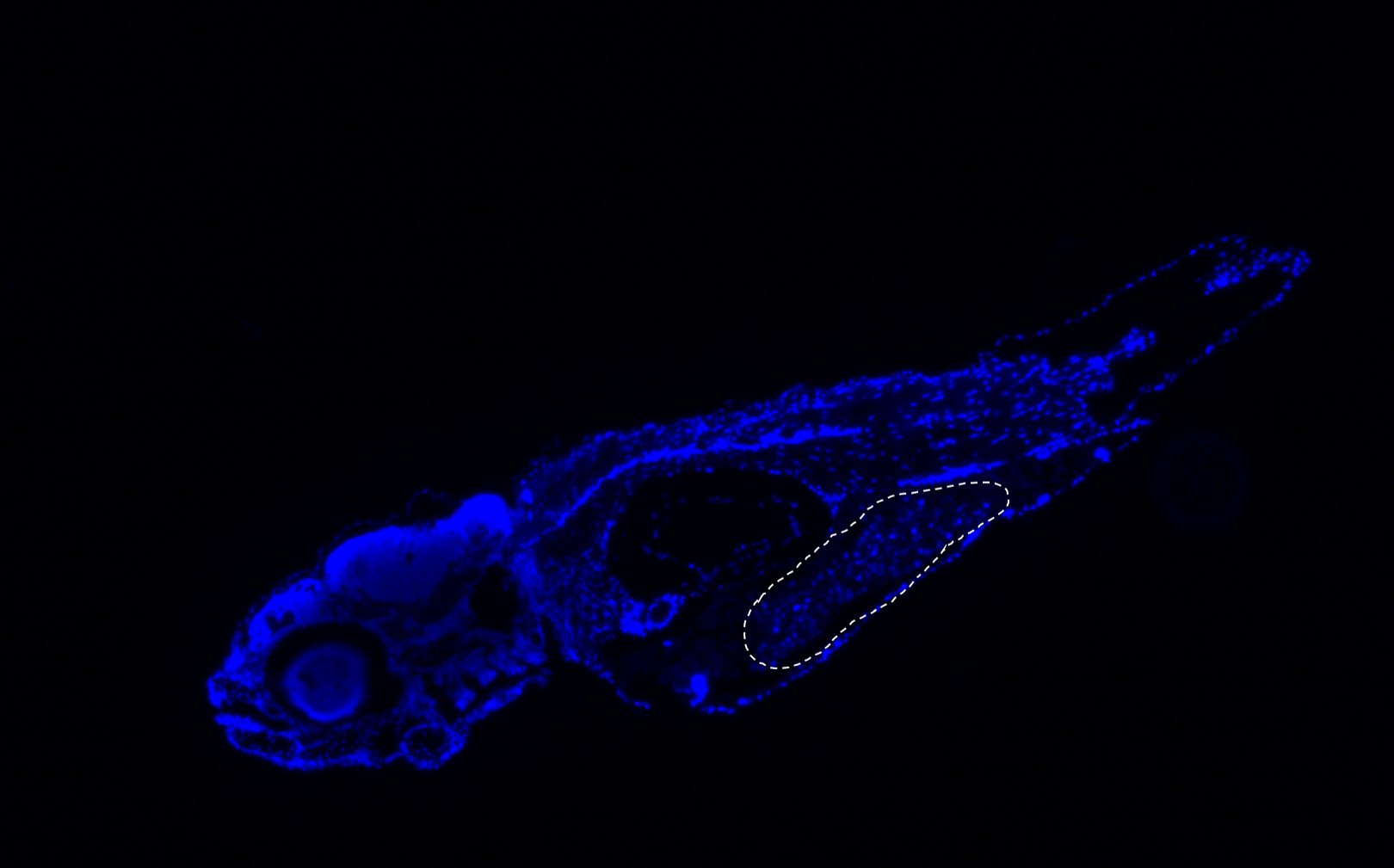
We established a zebrafish (Danio rerio) embryo human pediatric tumor cell xenograft model to investigate tumor growth and tumor progression (e.g., migration). This system is based on microinjection of pediatric (patient-derived) tumor cells into the yolk sac and perivitelline space (PVS). Moreover, we are additionally establishing an orthotopic pediatric brain tumor zebrafish model through microinjection of cells into zebrafish blastulas
Functional analysis of therapy resistance
Even though survival chances for children with malignant cancers have increased substantially over the last few decades, too many children still cannot be cured and succumb to disease. In many instances, this is either due to a lack of suitable treatment options in certain tumor entities or due to treatment resistance either existing before treatment or developing during therapy. The aim of our projects is the identification of actionable cellular alterations in pediatric nervous system tumors (neuroblastoma, medulloblastoma, glioma, ependymoma) via high-throughput drug screening.
In the case of treatment-resistant neuroblastomas, we are investigating how inhibition of cell surface growth receptors, drug efflux pumps and extracellular matrix components (e.g., MMPs) can sensitize resistant cells to chemotherapy. An additional focus of our studies is the lysosome, a cellular organelle responsible for the degradation of cellular macromolecules. A number of studies have suggested that lysosomes contribute to the chemoresistance of cancer cells by various mechanisms, including autophagy and lysosomal exocytosis. By systematically correlating high-throughput drug screens with high-content microscopy data, we are investigating how frequently lysosomal resistance mechanisms occur in pediatric nervous system tumors and whether they can be exploited as targets for combination therapies.
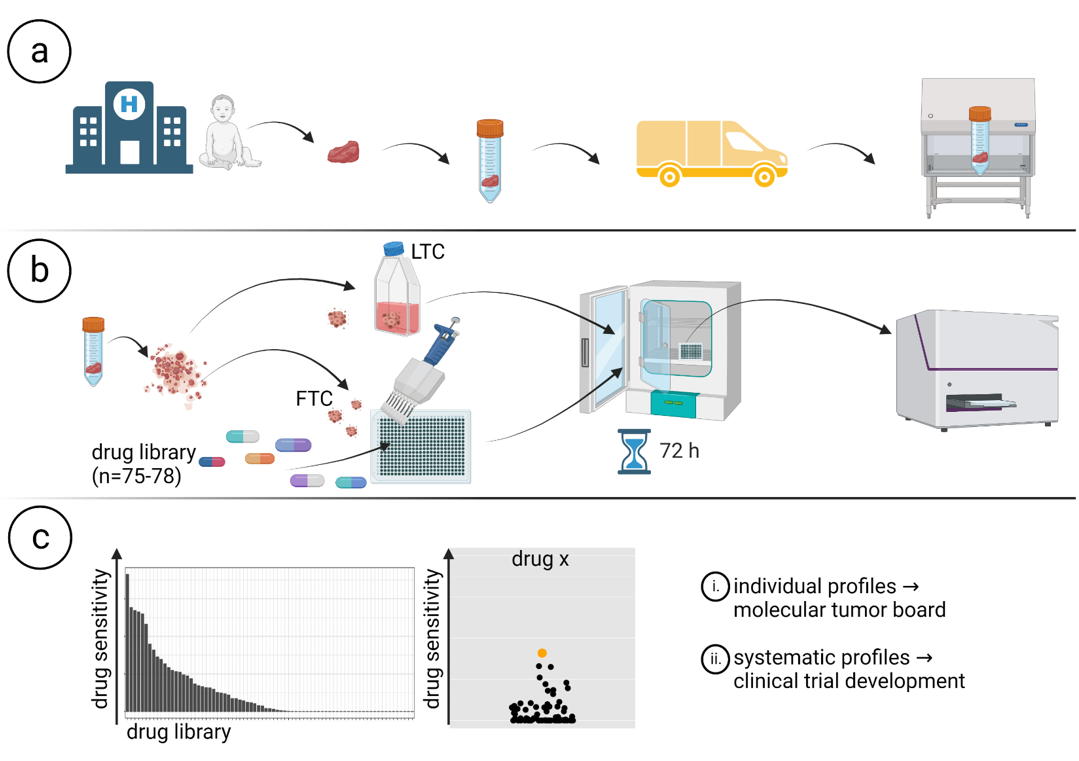
Workflow of INFORM personalized drug sensitivity profiling. Created with BioRender.com. (a) Sample collection and shipment. (b) Generation of patient-derived ex vivo fresh tissue spheroid cultures (FTCs) and treatment with a drug library. Readout: metabolic activity. (c) Data collection, analysis and preparation of drug sensitivity reports. Source: Peterziel et al.; npj PO 2022.
- Celikyürekli, Simay (Ph.D. student)
- Digel, Lea (student assistant)
- Friedenauer, Aileen (technical assistant)
- Gatzweiler, Charlotte (MD student)
- Gerloff, Xenia (student assistant)
- Herter, Sonja (Ph.D. student)
- Hugo, Anette (technical assistant)
- Körholz, Katharina (MD student)
- Loboda, Anna (Ph.D. student)
- Melero Emperador, Marta (Ph.D. student)
- Mohr, Jacqueline (student assistant)
- Müller, Michael (MD student)
- Najafi, Sara Dr. sc. hum. (Post-Doc)
- Oehme, Ina PD Dr. phil. nat. (group leader)
- Peterziel, Heike Dr. rer. nat. (staff scientist)
- Rösch, Lisa (Ph.D. student)
- Salem-Altintas, Rabia (MD student)
- Seiboldt, Till (MD student)
- Stroh-Dege, Alex (technical assistant)
- Weidmann, Marko (technical assistant)
- Zeiser, Constantia (MD student)