Spotlight - Group Westermann
Single-cell transcriptomic analyses provide insights into normal sympathetic system development
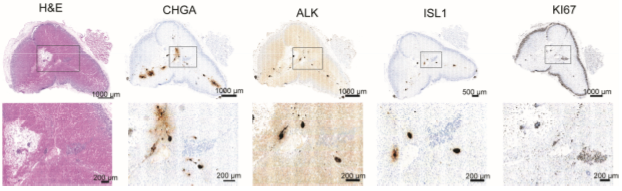
Studies on human adrenal gland development, which may shed light on the cellular neuroblastoma origin, are rare. We have initiated single-cell transcriptomics studies on the normal development of the human adrenal gland and the sympathetic ganglia in order to describe the normal counterparts and potential neuroblastoma cell(s)-of-origin. For this, we initiated a collaboration with the Human Developmental Biology Resource (HDBR, https://www.hdbr.org), which is a growing collection of human embryonic and fetal material ranging from postconception weeks 3 to 20. We used embryonal and fetal human tissues from the sympathetic nervous system and single-nuclei RNAseq (snRNA-seq) to characterize more than 200.000 cells (Jansky et al., 2021, Nature Genetics, 683-693). We specifically focused on the characterization of the medullary cells from the developing adrenal gland to understand the normal differentiation of sympathetic neurons and adrenal chromaffin cell lineages. The lineage relationships and differentiation dynamics of human adrenal medullary cell types during development were resolved. We identified transient cell populations such as bridge cells and connecting progenitor cells that are not found at later time points of normal development. Remarkably, we identified neural crest-derived SCPs to give rise not only to Schwann cells, but also adrenal chromaffin cells and proliferative neuroblasts.
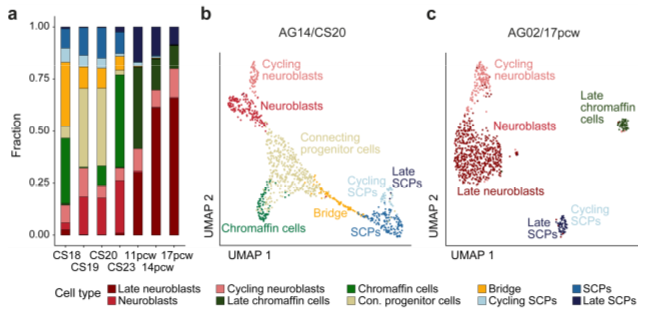
a, Bar plot depicting the proportion of adrenal medullary cell populations at each developmental time point. b, UMAP plot of adrenal medullary cells derived from one sample (AG14) at CS20. Cell populations are indicated by color and labels. c, UMAP projection of adrenal medullary cells of one sample (AG02) at 17 pcw with cell populations marked by color and labels. a-c, Adapted from (Jansky et al., 2021).
- PD Dr. Frank Westermann (Division Head)
- Dr. Kai-Oliver Henrich (Senior scientist)
- Dr. Sina Kreth (Senior scientist)
- Dr. Anand Mayakonda (Group leader, Bioinformatics)
- Dr. Sabine Stainczyk (Project manager)
- Cedar Schloo (PhD student)
- Pravin Velmurugan (PhD student)
- Ilayda Özel (PhD student)
- Dr. Michael Müller (MD/PhD student)
- Pascal Kohmann (MD student)
- Yeo-Eun Yi (MD student)
- Maximilia Eggle (MD student)
- Young-Gyu Park (Lab manager)
- Elisa Maria Wecht (Bioengineer)
- Manas Segal (Master student)
- Adrianna Podolak (Master student)
- Franziska Pohl (MD student)
- Claudia Müller (Lab technician)
- Marianne Riering (Master student)
- Yagmur Dölalan (Master student)